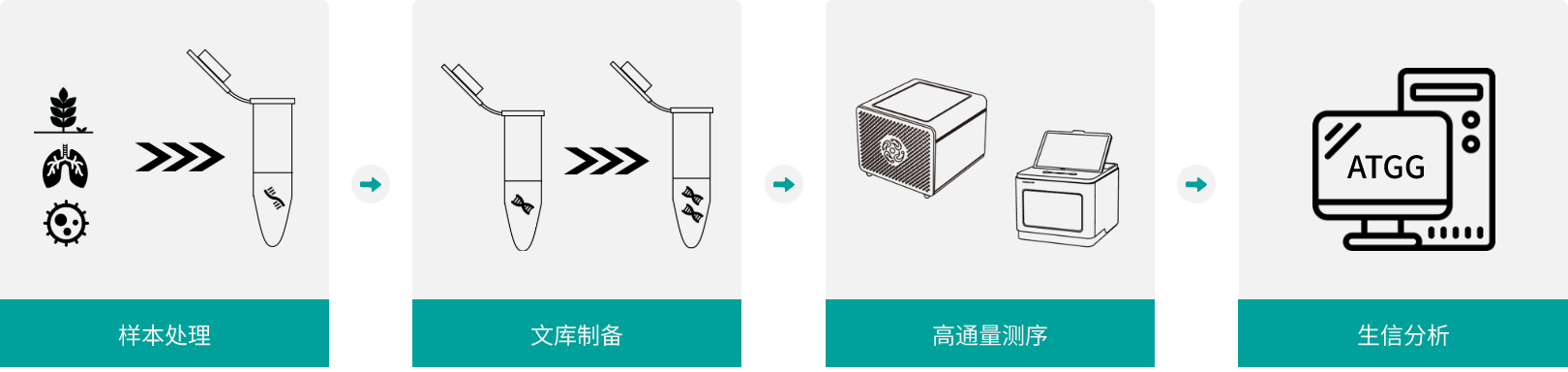
ATOPlex平台
——您身边的靶向测序专家
灵活设计,流程简便——提供从单个基因到10,000+靶点的高效经济型靶向测序解决方案,全面释放您的测序潜力
ATOPlex平台专注于靶向测序,提供高度灵活的靶向测序解决方案。平台依托超高重PCR扩增技术体系,快速提供端到端的整体测序解决方案。贯通基础科研到产业转化的全链条,深度赋能生命科学探索与医疗产业发展。
ATOPlex平台专注于靶向测序,提供高度灵活的靶向测序解决方案。平台依托超高重PCR扩增技术体系,快速提供端到端的整体测序解决方案。贯通基础科研到产业转化的全链条,深度赋能生命科学探索与医疗产业发展。
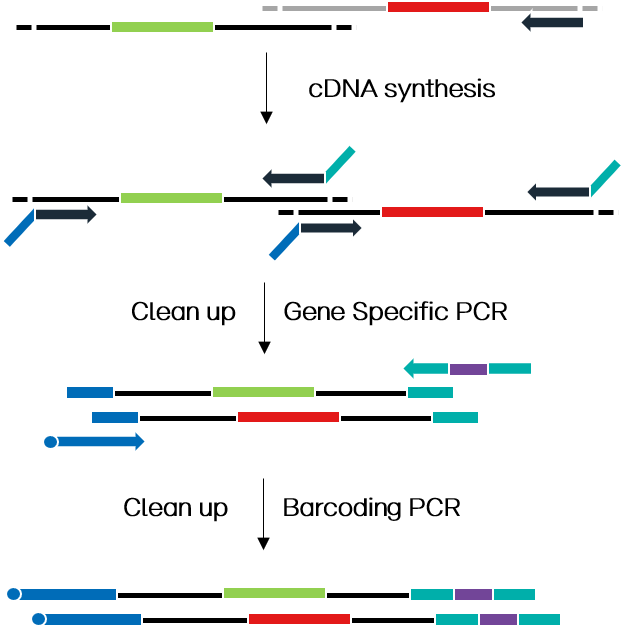
ATOPlex两轮PCR技术路线

图1:安捷伦2100电泳图显示出文库的高特异性
图2:Pa-SNP panel不同GC含量扩增子的扩增性能
图3:Pa-SNP Panel两次独立实验不同区域深度的稳定性